
A Modulator is a metabolite that, when bound to the allosteric site of an enzyme, alter its kinetic characteristics. The modulators of allosteric enzymes may be either stimulatory or inhibitory.
Many enzymes do not demonstrate hyperbolic saturation kinetics or typical Michaelis-Menten kinetics. Graphs of initial velocity vs. substrate demonstrate the sigmoidal dependence of V on S, much as we discussed with hemoglobin binding of dioxygen.

Enzymes that display this non-Michaelis-Menten behavior have common characteristics.
- These are multi-subunit
- It binds other ligands at sites other than the active site (allosteric sites)
- It can be either activated or inhibited by allosteric ligands
- It exists in two major conformational states, R and T
- These often control key reactions in major pathways, which must be regulated.
What are Allosteric Enzymes?
What is an Allosteric Enzyme? Jacob & Monod name such type of enzymes acts Allosteric enzymes. Allosteric means “Another site” (or) ” “Another space” (or) “Other than active site”.
Since allosteric enzymes possess an additional site other than a catalytic site to which a specific effector (or) modulator is reversibly and noncovalently bound.
These were named “Allosteric Enzymes“. These are the following are characteristics of allosteric enzymes
Salient Features of Allosteric Enzymes
What is allosteric regulation? The characteristics or salient features of allosteric enzymes are given below:
- Allosteric enzymes exist in two conformational states “R-state” and “T-state“.
- Non-allosteric enzymes show a rectangular hyperbola type of curve when Vo versus [S] was plotted. Whereas allosteric enzymes show the sigmoidal curve.
- Allosteric enzymes consist of more than one subunits i.e., they are oligomeric. These are usually much larger more complex having a high molecular weight and often more difficult to purify.
- Allosteric enzymes are subjected not only to the inhibition but also activated by modulators i.e., effectors can be positive (or) negative in their action.
- Allosteric enzymes catalyzed an irreversible reaction in a metabolic pathway.
- The reaction catalyzed by allosteric enzymes committed and rate-limiting steps.
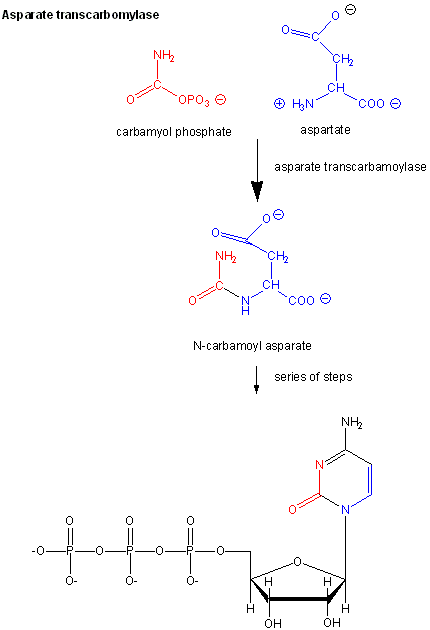
Examples of these enzymes include glycogen phosphorylase (the enzyme which breaks down intracellular glycogen reserves) and Aspartate transcarbamoylase (ATCase), which catalyzes the first step in the synthesis of pyrimidine nucleotides.
CTP is an allosteric inhibitor of this enzyme, which makes physiological sense since high levels of this pyrimidine nucleotide should inhibit the first enzyme in the synthesis of pyrimidines.
ATP is an allosteric activator. This also makes sense since if high levels of the purine nucleotide ATP are present, one would also want to balance the level of pyrimidine nucleotides.
Earlier we saw that cooperative binding equilibrium could be modeled with the Hill Equation, which was introduced into the equation
Y=Ln/(Kd+Ln)=Ln/(P50n+Ln)
where n is the co-operativity or Hill Coefficient. Likewise, for an enzyme which demonstrates cooperative (sigmoidal) initial rates plots,
vo=VmSnK0.50n+Ln
When n=1, the equation reduces to the classical hyperbolic Michaelis Menten equation. For values of n>1, sigmoidal plots are observed.
We found a more easily understandable molecular interpretation of the cooperative binding of oxygen to hemoglobin using the MWC model (T and R states).
- Diagnostic Enzymes: Enzymes in the Diagnosis of Pathology
- What are the Enzymes Involved in DNA Replication?
- Isoenzymes are Special Proteins with Catalytic activity. Why? (MCQ)
The MWC model has also been applied successfully to multi-subunit enzymes which display cooperative, sigmoidal kinetics. In this model, allosteric inhibitors (which often don’t resemble the substrate) bind preferentially to the T state, leading to lower activity.
While allosteric activators bind preferentially to the R state, leading to greater activity. Activators shift the vo vs. S curve to the left while inhibitors shift it to the right (much like protons and carbon dioxide in hemoglobin binding). These allosteric ligands induce their effects by shifting the To⇌Ro equilibrium.
- What are the allosteric enzymes in glycolysis? Hexokinase, PhosphoFructokinase, and Pyruvate kinase are the best examples of Allosteric enzymes. They are showing virtually irreversible reactions.
- Phosphofructokinase is an allosteric enzyme that catalyzes Fructose-6-Phosphate to Fructose-1,6-bisphosphate, early step in the Glycolysis.
Control of Allosteric Enzymes
How do most cells regulate the activity of enzymes? Allosteric enzymes show two different types of control mechanisms. On the basis of which these are classified into two types.
- Heterotropic Enzymes: Which are stimulated (or) inhibited by an effector (or) modulator molecule other than their substrates. Eg: Threonine dehydratase (Substrate: Threonine, Modulator: Isoleucine).
- Homotropic Enzymes: Here substrate functions as the modulator. These enzymes contain two or more binding sites for the substrate. Eg: ATCase.
How Do Allosteric Effectors Change Vm and Km?
How does a noncompetitive inhibitor reduce an enzyme’s activity? We have just studied how competitive, uncompetitive, and non-competitive (or mixed) inhibitors influence the apparent Km and Vm values for enzymes that display Michaelis-Menten kinetics.
How are Vm and Km influenced by allosteric enzymes? The example given above (ATCase), analogous to effects observed in hemoglobin: oxygen binding, influence the apparent Km, but not the Vm.
Remember that in the case of hemoglobin binding curves, the allosteric activators and effectors we discussed shifted the sigmoidal binding curves to the left or right, but all reached a plateau at the same fractional saturation value of 1.
Allosteric enzyme systems that behave like this are called K systems. Enzymes in which allosteric regulators change Vm, called V systems, are also known.
V systems display hyperbolic vo vs S curves in which activators display a greater apparent Vm and inhibitors display a lower Vm without affecting the apparent Km.
In these systems, both the T and R forms have the same affinity for substrate (hence the same apparent Km).
This would be analogous to a situation in the MWC model where KR/KT =1 which also gave hyperbolic, not sigmoidal Y vs. curves.
This difference in V systems is that the R and T states have different catalytic rate constants, kcat, for turnover of the bound substrate (hence different apparent Vmax values).
In addition, the activator A and inhibitor I bind to the R and T forms with different affinity, which again shifts the To⇌Ro equilibrium in the presence of the allosteric effectors.
Cooperative binding of dioxygen to hemoglobin, regulated by allosteric effectors (protons and carbon dioxide), was ideal for an oxygen transport system that must load and unload oxygen over a narrow range of oxygen concentrations and allosteric effectors.
Allosteric enzymes are usually positioned at key metabolic steps that can be regulated to activate or inhibit whole pathways.
Biological Significance of ATCase Regulation
- Both purines & Pyrimidines are essential for Nucleic acid synthesis. Activation by ATP signals that energy is available for DNA replication and leads to the synthesis of needed pyrimidines
- Feedback inhibition by CTP indicates that N-carbamoyl aspartate and subsequent intermediates in the pathway are useless when pyrimidines are abundant.
- ATCase consists of separable catalytic and regulatory subunits. The regulatory property of ATCase is identified by treating with para-hydroxy mercury benzoate. That specifically destruct the regulatory site without affecting the catalytic site.
- The catalytic and regulatory subunits of ATCase are composed of discrete catalytic (C) and regulatory sites (R). C & R are interacted with the native enzyme to produce its allosteric behavior. In a native enzyme molecule, there are three “C-subunits” and two “R-subunits”. The three C-subunits are larger in size and are catalytically active, but no response to ATP and CTP. The smaller subunit R is catalytically inactive but can bind with CTP or ATP and it consists of two R-chains.
