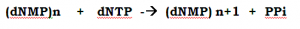The first postulate the central dogma, experimentally proved and is that the DNA is capable of self-replication. This was immediately deduced by Watson and Crick that each DNA strand uniquely specifies its complement, but it took longer for the details of the mechanism are elucidated.

The first question was that of the conservation of the replication mechanism.
The now-classic experiments of Meselson and Stahl proved that DNA replication is semi-conservative, in that each of the sister molecules inherits one strand of the parental DNA.
It would have been perfectly possible to imagine a conservative mechanism, where the two strands of the molecule daughter have been synthesized de-novo, using parental strand as a template.
The DNA replication mechanism is catalyzed by the groups of enzymes. In the group of enzymes, DNA Polymerases are the major catalytic proteins with polymerization property by using Nucleotides like ATP, TTP, CTP, and GTP (not UTP).
DNA Polymerases have the same function in both Prokaryotes and Eukaryotes but it has a difference in their structure.
So, in this topic (or) article we are providing the complete material on DNA Polymerases enzyme structure and functions in both Prokaryotes and Eukaryotes.
DNA Polymerases
Enzymes catalyzing DNA synthesis on a DNA template are DNA Polymerases. They perform two primary functions in the cell: the synthesis of DNA during genome replication, and the re-synthesis of missing DNA following damage of recombination, and following primer excision from the lagging strand.
In both prokaryotes and eukaryotes, specialized DNA polymerases are dedicated to replication and repair functions, the former sometimes being termed DNA replicases. All DNA polymerases possess a 5′->3′ polymerase activity. and pyrophosphorolysis activity, which together facilitates DNA synthesis.
- What is the Fidelity of DNA Replication in Normal?
- What is the origin of replication in Prokaryotes and Eukaryotes
Prokaryotic DNA Polymerases
In 1957, “Arthur Korenberg” showed that extracts of E.Coli contain a DNA polymerase (now called Polymerase I or Pol I ).
This enzyme able to synthesize DNA from four precursor molecules, namely the four deoxynucleotides 5’-Phosphate (dNTP), dATP, dGTP, dCTP and dTTP, as long as a DNA molecule to be copied (a template DNA) is provided.
Neither 5’-monophosphates nor 5’-diphosphates, nor 3’-(mono-, di-, or tri-) phosphates can be polymerized only the 5’-triphosphates are substrates for the polymerization reaction.
E. Coli has at least Three DNA polymerases:
- DNA Polymerases-I
- DNA Polymerases-II
- DNA Polymerases-III
1) DNA Polymerases-I
DNA polymerase I in prokaryotes is far from irrelevant, however. This enzyme serves as a host of “Clean-up” functions during replication, recombination, and repair.
These special functions are enhanced by an additional enzymatic activity of DNA polymerase I, a 5’->3’ exonuclease activity.
This activity is distinct from the 3’->5’ proofreading exonuclease and is located in a distinct structural domain that can be separated from the enzyme by mild protease treatment.
When the new Okazaki fragment is complete, the RNA primer is removed by DNA polymerase-I and is replaced with DNA by the sea enzyme.
When the 5’->3’-exonuclease domain is removed, the remaining fragment (M.w.-68,000) retains the polymerization and proofreading activities and is called the “Large (or) Klenow fragment”.
This Klenow fragment lacks the “5’->3’-exonuclease activity”. The structure of the Klenow fragment has been demonstrated, and it is this fragment of DNA polymerase I, the 5’->3’ exonuclease activity of intact DNA polymerase I permit it to extend DNA strand even if the template is already paired to an existing strand of nucleic acid.
Using this activity, DNA polymerase I can degrade (or) displace a segment of DNA (or RNA) paired to the template and replace a segment of DNA (or RNA) paired to the template and replace it with newly synthesized DNA.
Most other DNA polymerizes including DNA polymerase III, lack a 5’->3’ exonuclease activity.
Fundamental Reaction:
The fundamental reaction is a ‘Nucleophilic attack’ by the 3’-hydroxyl group of the nucleotide at the 3’ end of the growing strand on the 5’-a-phosphorous of the incoming deoxynucleoside 5’-triphosphate.
Inorganic phosphate is released in the reaction.
2) DNA polymerase II
DNA polymerase II is a minor component of the cell during normal growth but is inducible by the SOS response. It appears that this enzyme allows nucleotide incorporation opposite AP sites. DNA polymerase II appears to have a highly specialized DNA repair function.
This enzyme participates in “Base-Excision repair” and “ Nucleotide-Excision repair”
Base-Excision repair:
- Every cell has a class of enzymes called “DNA glycosylases” that recognize particularly common DNA lesions and remove the affected base by cleaving the N-glycosyl bond.
- This creates an “Apurinic” (or) “Apyrimidinic site” in the DNA, both commonly referred to as a “basic” (or) “AP sites”.
- Each DNA glycosylase is generally specific for one type of lesion.
Nucleotide-Excision repair
- DNA lesions that cause large distortions in the helical structure of DNA generally are repaired by the nucleotide-excision system.
- In E.Coli the key enzyme is made up of THREE subunits, products of the “uvr.A, uvr-B, uvr-C genes”, and is called the “ABC Exonuclease”.
- This enzyme recognizes many types of lesions, including “Cyclobutane-pyrimidine Dimers”, “6,4-photoproducts” and several other types of “base additions”.
- The term “Exonuclease” is meant to distinguish this activity from that of standard endonucleases.
3) DNA Polymerases-III
Prokaryotic DNA Polymerase-III is a very complex enzyme. In its most active form it is associated with nine (or) more other proteins to form the “Pol III HOLOENZYME”, occasionally termed Pol III.
The term holoenzyme refers to an enzyme that contains several different subunits and retains some activity even when one (or) more subunits is missing.
The smallest aggregate having enzymatic activity is called the “CORE ENZYME”. The activity of the core enzyme and the holoenzyme are usually very different.
DNA polymerase III has a highly complex protein composed of 10 different polypeptides.
Overall, the enzyme has an “Asymmetric dimeric structure”. It contains two copies of most subunits and two catalytic sites for nucleotide addition.
Prokaryotic DNA polymerase III:
DNA polymerase III is the principle replicative DNA polymerase of E.Coli. it is a multisubunit complex. The holoenzyme (Apoenzyme [protein part] + Coenzyme = Holoenzyme) functions as a “Heterodimer” of complexes at the replication fork, with each monomer seeing to the synthesis of one daughter strand.
α + ε + Φ – subunits -> Core enzyme
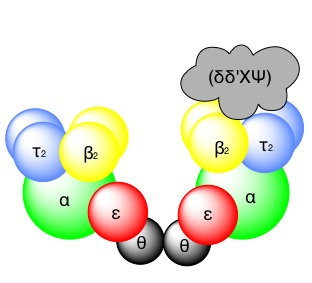
The core enzyme, which contains the essential enzyme activities. The assembly of the holoenzyme in vivo occurs as follows: the beta-subunit functions as a dimer and forms a ring (or) clamp, which can slide along single-stranded DNA.
The beta-subunit is located onto template-primer by the g-complex, an ATP-dependent process, to form the “pre-initiation complex”.
The loading of the beta-subunit allows the core –enzyme to bind, and the addition of the t-subunit facilitates dimerization.
The holoenzyme is “Symmetrical” except for the g-complex, which is associated with only one of the monomers. The g-complex is required for both loading and unloading the beta-subunit from DNA.
The presence of the g-complex allows the beta-subunit to dissociate from the template-primer when the polymerization encounters the 5’end of previously synthesized Okazaki fragments on the retrograde template.
Subunits of the E.Coli DNA polymerase III holoenzyme and their proposed functions
| Subunit | Gene | Properties and proposed function |
| Core Subunits α | dna.E (pol.C) | 5’–> 3’ polymerase activity, required for DNA synthesis |
| ε
| dna .Q (mut.D) | 3’–>5’ exonuclease activity required for proofreading. |
| ø | Unassigned | Function uncertain may help to assemble other subunits. |
Accessory subunits (₸) | dna.Xa | DNA dependent ATPase, required for initiation. Promotes dimerization |
| γ | dna.Xa | Associates with four peptides to form a DNA dependent ATPase known as the gamma-complex required for initiation, facilitates beta-subunit binding. |
| δ, δ’,Χ, Ψ
| Unassigned | Associate with “ g “ to form the g-complex. |
| β | dna.N | “Sliding clamp”, which increases the processivity of the holoenzyme. ‘beta’ binds to DNA to form a precipitation complex, a process which requires the ATP-dependant activity of the gamma-complex dna.N is induced by the “SOS response” |
Eukaryotic DNA Polymerase
Eukaryotic cells contain FOUR nuclear DNA polymerases and a fifth, which is responsible for organelle genome replication. The nuclear enzymes are DNA polymerases α, β, δ, and ε DNA polymerase alpha and delta: α δ
- DNA polymerase alpha and delta are responsible for chromosomal replication.
- DNA polymerase delta has a proofreading capability.
- DNA polymerase d binds an accessory factor called “Proliferating cell nuclear antigen (PCNA), a cyclin analogous to the E.Coli polymerase III beta subunit in that it acts as a sliding ring to increase enzyme processivity.
- delta Synthesizes both the leading and lagging strands.
- To the lagging strand, synthesis provides the template for an accessory factor, “Replication factor.C (RF.C)”, this role analogous to that of the E.Coli gamma-complex.
- DNA polymerase associated with “Primase” activity but no 3’–>5’ exonuclease activity, whereas DNA pol delta has a “Proofreading capability”.
DNA polymerase Epsilon
- The role is unclear.
- It is structurally very similar to DNA pol delta but does not associate with PCNA.
- It may be involved in DNA repair, like DNA polymerase beta
DNA polymerase gamma:
- Responsible for the replication of mitochondrial DNA and a similar enzyme has been isolated from plant chloroplast.
Functions of DNA polymerase subunits:
- Eukaryotic DNA polymerase alpha -> Lagging strand priming
- Eukaryotic DNA polymerase beta -> Repair polymerase
- Eukaryotic DNA polymerase gamma -> Organelle polymerase
- Eukaryotic DNA polymerase delta -> Principle replicative polymerase
- Eukaryotic DNA polymerase epsilon -> Unknown
Differences between Eukaryotic DNA Polymerase vs Prokaryotic DNA Polymerase
Prokaryotes
- DNA pol III is involved in invivo replication of DNA.
- DNA pol I is the repair enzyme.
- Primase synthesizes primer in EColi.
Eukaryotes
- DNA pol α and delta are involved in invio replication of nuclear DNA whereas DNA pol gamma for mitochondrial DNA.
- DNA pol beta and epsilon function as DNA repair enzymes.
- DNA pol alpha synthesizes primers for both leading and lagging strands