
Ramachandran plot is a plot of the torsional angles – phi (φ)and psi (ψ) – of the residues (amino acids) contained in a peptide. In sequence order, φ is the N(i-1),C(i),Ca(i),N(i) torsion angle and ψ is the C(i),Ca(i),N(i),C(i+1) torsion angle. The plot was developed in 1963 by G. N. Ramachandran, by plotting the φ values on the x-axis and the ψ values on the y-axis, as for the image at left. Plotting the torsional angles in this way graphically shows which combination of angles is possible.
What Is Peptide Linkage? Joining of an α-carboxyl group of one amino acid to the α-group of another. Formation of the bond involves loss of water molecule from the two amino acids, the bond can be broken by hydrolysis.
Peptide Bond Is Planar Because the torsional angles of each residue in a peptide define the geometry of its attachment to its two adjacent residues by positioning its planar peptide bond relative to the two adjacent planar peptide bonds, thereby the torsional angles determine the conformation of the residues and the peptide. Many of the angle combinations, and therefore the conformations of residues, are not possible because of steric hindrance. By making a Ramachandran plot, protein structural scientists can determine which torsional angles are permitted and can obtain insight into the structure of peptides. The scene on the right is the Ramachandran plot of ribonuclease H.
Who is G N Ramachandran?
Gopalasamudram Narayana Ramachandran (8 October 1922 – 7 April 2001) is an Indian biophysicist and crystallographer who, along with Gopinath Kartha, worked out the triple helical structure of collagen.
G N Ramachandran is an Indian biophysicist who was known for his work that led to his creation of the Ramachandran plot for understanding peptide structure. He was the first to propose a triple-helical model for the structure of collagen. He also made other major contributions in biology and physics.
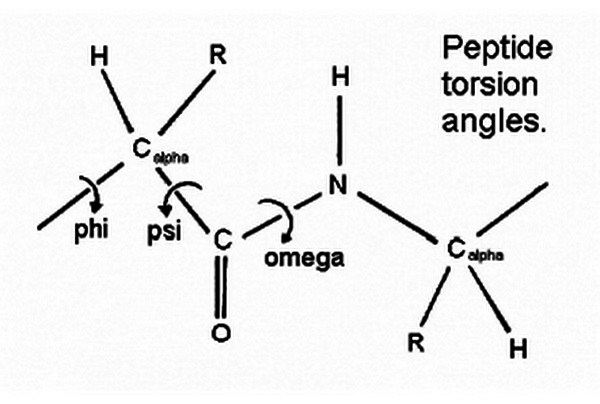
Ramachandran Plot and Peptide Torsion Angles
The figure below shows the three main chain torsion angles of a polypeptide. These are phi (ψ), psi (φ), and omega (w).
Secondary structure plot regions
Secondary structures of a peptide are segments of the peptide that have ordered and repetitive structure, and the repetitive structure is due to a repetitive confirmation of the residues and, ultimately, repetitive values of ψ and φ. The different secondary structures can be distinguished by their range of ψ and φ values with the values of different secondary structures mapping to different regions of the Ramachandran plot. Two common examples of secondary structure are illustrated below.
α-helix
The scene on the right shows the axis of the α-helix rotating in the y-plane. When viewing the helix on end, observe the open center of the helix. Planes are drawn on some of the peptide bonds to emphasize that in an α-helix the planar peptide bonds rotate about the axis of the helix.
The Ramachandran plot of this peptide has points clustered about the values of ψ= -47o and φ= -57o which are the average values for α-helices.
Adding the values of two other helical segments demonstrates that data from all three appear in one large cluster and that the helical segments cannot be distinguished by the differences in their ψ and φ values.
β-sheets
Display a two segment twisted β-sheets. Draw planes of the peptide bonds. Most β-sheets in globular proteins are twisted sheets which do not have even parallel pleads.
Closer view of β-sheets. The Ramachandran plot of this twisted sheet has points clustered about the values of ψ= -135o and φ= +135o which are the average values for twisted sheets.
Adding the values of three other sheet segments more clearly defines the area in which values for twisted sheets are located.
Plot regions limited by Steric hindrance
Most combinations of ψ and φ are sterically forbidden, as illustrated in the tripeptide, Glu-Ser-Ala. With Ser having values of ψ = -116o and φ = 55o the Ser side chain is in contact with Ala, colored blue.
In plots of native peptides, the data points will form clusters in the several areas in which steric hindrance does not occur.
These regions are illustrated in the Figure on the left. The core regions contain the most favorable combinations of ψ and φ and contain the greatest number of points.
Plots of some proteins contain a small third core region in the upper right quadrant. The allowed regions (green in the Figure) can be located around the core regions or can be unassociated with a core region, but they contain fewer data points than the core regions.
- What is DNA Polymerase and its function in DNA Replication
- What is Fidelity of DNA Replication in Normal?
The generous regions (not shown in the Figure)extend beyond the allowed regions. The remaining areas are the disallowed regions. Observe that the data point – 116o and +55o for Ser of the above tripeptide would fall in the lower right quadrant which contains only a disallowed region. If the Serine in the tripeptide has ψ and φ values of -47o and -57o which are in the core region in the lower left, the Serine side chain is rotated away from Ala and is not in contact with Ala. Most, if not all, of the points in the above plots for α-helix and β-sheet, are in one of the core areas.
Glycine
Since Gly has only a hydrogen as a side chain, steric hindrance is not as likely to occur as ψ and φ are rotated through a series of values. The tripeptide Glu-Gly-Ala with Gly having ψ and φ values of -116o and +55o respectively, does not show the steric hindrance that the Glu-Ser-Ala had. For that reason, Gly will frequently plot in the disallowed region of a Ramachandran plot. Nearly all of the data points in the disallowed region in the above Figure are Glycine points.
Functionally relevant residues
Functionally relevant residues may occasionally have torsion angles that plot to the disallowed regions of a Ramachandran plot. The specific geometry of these functionally relevant residues, while energetically unfavorable, may be important for the protein’s function, catalytic or otherwise.
Ramachandran Plot Explanation
(Smart Notes Description)
How to read Ramachandran plot? A Ramachandran plot is a way to visualize backbone dihedral angles ψ against φ of amino acid residues in protein structure. A Ramachandran plot can be used in two somewhat different ways. One is to show in theory which values, or conformations, of the ψ and φ angles, are possible for an amino-acid residue in a protein. A second is to show the empirical distribution of data points observed in a single structure.
Ramachandran plot analysis
The coloring/shading on the plot represents the different regions described in Morris et al. (1992): the darkest areas correspond to the “core” regions representing the most favorable combinations of phi-psi values. Ideally, one would hope to have over 90% of the residues in these “core” regions. The percentage of residues in the “core” regions is one of the better guides to stereochemical quality. The different regions were taken from the observed phi-psi distribution for 121,870 residues from 463 known X-ray protein structures.
The two most favored regions are the “core” and “allowed” regions which correspond to 10° x 10° pixels having more than 100 and 8 residues in them, respectively. The “generous” regions were defined by Morris et al. (1992) by extending out by 20° (two pixels) all around the “allowed” regions. In fact, the authors found very few residues in these “generous” regions, so they can probably be treated much like the “disallowed” region and any residues in them investigated more closely.
Final Words
The nature of the covalent bonds in the polypeptide chain places constraints on the structure. The peptide bond exhibits partial double bond character that keeps the entire peptide group in a rigid planar configuration. The N-Cα and Cα-C bonds can rotate with bond angles Φ and Ψ, respectively. Secondary structure can be defined completely if the Φ and Ψ angles are known for all amino acid residues in that polypeptide segment. This Ramachandran plot explains the nature and rotational angles if Peptide bonds in Polypeptide chains.
