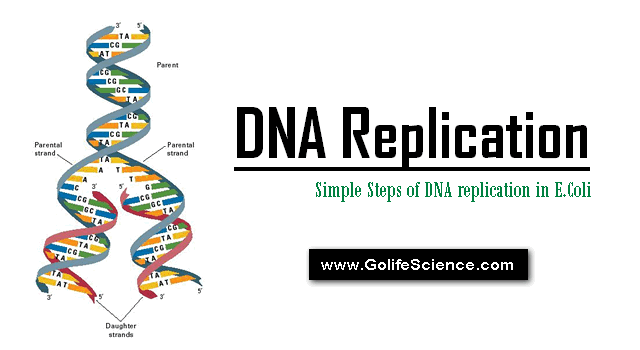
DNA replication steps start at unique chromosomal origins, processed bidirectionally, and is semi-conservative. DNA replication mechanisms depend on prior events: identification of a system capable of sustaining in-vitro replication of small plasmids carrying “Ori.C”

DNA Replication Definition:
DNA replication is the process by which a double-stranded DNA molecule is copied to produce two identical DNA molecules. Replication is an essential process because, whenever a cell divides, the two new daughter cells must contain the same genetic information, or DNA, like the parent cell.
Basic Steps of DNA Replication
The synthesis of a DNA molecule (or) DNA Replication process can be divided into THREE stages:
- Initiation
- Elongation
- Termination
1. Initiation Stage
The E.Coli DNA replication process, called “Ori.C”, consists of 245 base pairs, many of which are highly conserved among bacteria.
The key sequences for this discussion are two series of short repeats; there repeats of a 13 base pair sequence and four repeats of a 9 base pair sequence.

At least “Eight different enzymes” (or) “Proteins” participate in the initiation phase of replication. They open the DNA helix at the origin and establish a preparing complex that sets the stage for subsequent reactions.
The key component in the initiation process is the Dna.A protein. A complex of about twenty Dna.A protein molecule binds to the four Nine base pairs repeats in the origin.
In a reaction that requires ATP and is facilitated by the bacterial histone-like protein “HU”, the Dna.A protein recognizes and successfully denatures the DNA in the region of the there 13 base pairs repeats, which are rich in A=T pair.
The Dna.B protein then binds to this region in a reaction that requires the Dna.C protein. Dna.B is a “Helicase” that unwinds duplex DNA. The helicases constitute a class of enzymes that can move along a DNA duplex utilizing a class of enzymes that can move along a DNA duplex utilizing the energy of ATP hydrolysis to separate the strands.

The separated strands are inhibited from subsequently re-annealing by the E.Coli Single-Stranded binding protein (SSBP), which binds to both separated strands. Multiple molecules of SSB bind cooperatively to single-stranded DNA, stabilizing the separated DNA strands and preventing renaturation.
DNA replication must occur only once in each cell cycle. Initiation is the only phase of replication that is regulated, but the mechanism is not yet well understood.
The Dna.A protein hydrolyzes its tightly bound ATP slowly (about 1 hour) to form an inactive Dna.A–ADP complex. Reactivating this complex is facilitated by an interaction between Dna.A protein and acidic phospholipids in the bacterial plasma membrane.
Initiation at inappropriate times is prevented by the presence of the inactive Dna.A-ADP complex by the binding of a protein called “ICI” (Inhibitor of chromosomal initiation) to the 13 base pair repeats, and perhaps by other factors.
2. Elongation Stage

The elongation phase of replication consists of two seemingly similar operations that are mechanically quite distinct: Leading strand synthesis and Lagging synthesis. Several enzymes at the replication fork are important to the synthesis of both strands. “DNA helicases” unwind the parental DNA. “DNA topoisomerases” relieve the tropological stress included by the “Helicases”, and “SSBP” stabilizes the separated strands.
a) DNA Replication leading strand synthesis
- It begins with the synthesis by “Primase” of a short (10 to 60 nucleotide) RNA primer at the replication origin.
- Deoxyribonucleotides are then added to this primer by DNA polymerase-III once was begun, leading strand synthesis proceeds continuously, keeping pace with the replication fork.
b) DNA Replication Lagging strand synthesis

It must be accomplished in short fragments (Okazaki fragments) synthesized in the direction opposite to the fork movement.
- Each fragment must have its own RNA primer synthesized by “primase”, and positioning of the primers must be controlled and coordinated with fork movement.
- The regulatory apparatus for “lagging strand synthesis” is a traveling protein machine called a “ PRIMOSOME”, which consists of several different proteins including the Dna.B protein, Dna.C protein, and Primase.

The primosome moves along the lagging strand template in the 5’à3’ direction, keeping pace with the replication fork.
As it moves the primosome at intervals compels primase to synthesize short (10 to 60) residues. RNA primer to which DNA is then added by DNA polymerase-III.
The direction of the synthetic reactions of “Primase” and “Polymerase-III” is opposite to the direction of the primosome movement.
When the new Okazaki fragment is complete, the RNA primer is removed by DNA polymerase-I and is replaced with DNA by the same enzyme. The remaining nick is sealed by “DNA ligase”.

3. Termination Stage
a) Termination of Replication of a circle
There are two possible modes of replication:
- There are defined termination sequences, or
- Two growing points collide and termination occurs whenever the collision point happens to be.
In both cases, termination might occur exactly halfway around the circle. Both termination modes have been observed.
Termination has a “Topological problem”. When double-stranded circular DNA replicates semi conservatively, the result is a pair of circles that are linked as in a chain.
Such a structure is called “CATENANE”. Catenated molecules have been observed in numerous systems, and evidence is accumulating to indicate that they result from replication.

“DNA Gyrase” is capable of de-catenating two circles, which enzyme is responsible for the separation of daughter molecules.E.coli chromosome and several plasmids carry specific sequences, called “ter sites”, where TBP ( ter binding protein) or ‘Tus protein’ bind.
In the termination zone of E.coli, there are three ter sites (ter A, ter D and terE) for a counter-clockwise fork. These six sites are arranged in an overlapping manner, laving no “Replication – free” gap on the chromosome. TBP-ter complexes formed at “ter” sites stall the replication fork, by inhibiting the DNA helicase or DnaB.
When this termination zone is deleted, replication stops, simply by the meeting of opposite replication forks, suggesting that the termination zone is not essential.
b) Termination in ‘Linear’ DNA molecules:
E.Coli phage T7 DNA replicates as a linear molecule. The origin is located 17% of the total distance from the left end of the molecule and replication is bidirectional. Initially, there is a single replication bubble (molecule-I), and when the leftward fork reaches the terminus, the molecules assume a “Y”-form (molecule-III).
“Ribonucleases” exist that can remove this RNA but once it has been removed, either a 5’-OH group would remain at the ends of the molecule.
Interaction of “Tus protein” with termination sites stops DNA replication:
- Although much research attention has been focused on replication origins, E.Coli DNA also has been shown to contain termination (TER) sites, which bind a specific protein called Tus.
- This protein may act to stop replication by preventing “Helicase (DnaB) from unwinding duplex DNA, thereby interrupting the function of the growing fork.
- The details of the mechanism that arrests the growing fork are not yet clear. Replication of the circular E.coli chromosome produces two interlocking daughter chromosomes, which must be separated.
DNA Replication Video
MCQ on DNA Replication (Check your Basics)
[mtouchquiz 6]


